
After construction, you can check the reliability of the result by Bootstrapping. You can construct the tree by these two methods- UPGMA and Neighbourhood Joining method.CLC Main Workbench phylogenetic tree editor allows you to have- circular and radial layouts, data import in CSV format, a tabular representation of metadata, collapsing nodes based on bootstrap values, re-arranging the nodes, visualization of metadata, coloring, and labeling of trees, curved edges, editable nodes, and line size, etc.The phylogenetic tree construction makes the understanding of such relations easier. This is a key aspect in understanding the ancient genetic relations between different groups of organisms. A great way to realize the evolutionary change that might have occurred. Multiple sequences can be aligned too to identify the mutations caused over time.You can even perform the pairwise comparisons, create comparison tables too.Join the alignments after you have edited them.It lets you remove changes, adjust the gaps, and edit the fixpoints. It is a powerful tool for editing alignments.
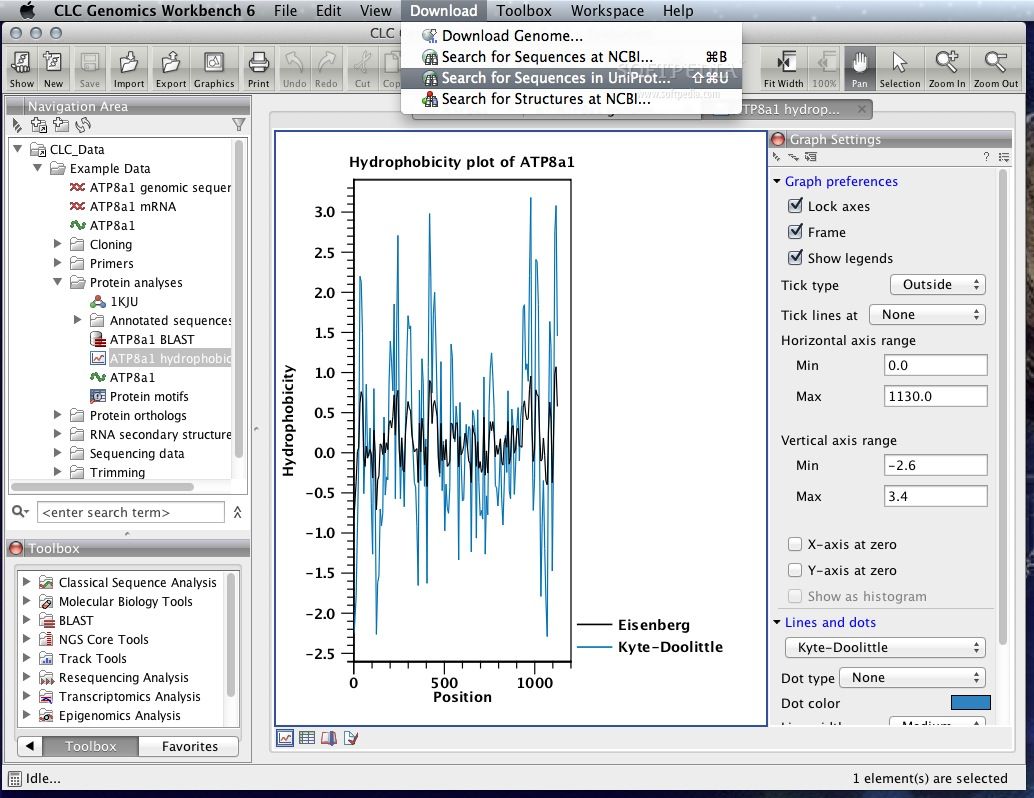
The realignment option lets you create alignment again.You can add gaps, delete gaps, copy annotations to other sequences, etc. The extra choice of editing the alignments is provided here. Just like any other sequence alignment tool, it displays the aligned regions and gaps.Alignments can be viewed via Alignment info and Nucleotide info options.One of them is fast and less accurate, while the other is slow and very accurate. CLC Main Workbench utilizes two algorithms for finding the alignments.This alignment algorithm has three parameters for calculating the gap costs. Image source: QIAGEN CLC Main Workbench Sequence AlignmentĬLC Main Workbench is efficient in aligning the nucleotides and proteins by using the Progressive alignment algorithms. Given below is a biomolecule structure generated from PDB 2R9R. A file with information on 3D Coordinates only can be used to generate entire structures. The “ Generate Biomolecule” option lets you generate biomolecules structures in CLC Main Workbench.The Protein structures can be aligned too.The transfer of annotation between the sequence and structure is feasible too. They link the sequence alignments to the molecule structure. There are some tools present in the 3D viewer that helps in linking the sequence and the structure.You can BLAST search the structure file against the PDB database.The visualization settings include- hydrogens, fog, clipping plane, 3D projection, coloring, etc. The visualization can be customized with different colors and styles of representation.


 0 kommentar(er)
0 kommentar(er)
